Taxonomy Development Tools User Interface Guide
Welcome to the Taxonomy Development Tools User Interface Guide. This document is designed to provide comprehensive details on navigating and utilizing the TDT interface efficiently. Whether you are looking to manage data tables, edit information, or leverage advanced features, this guide will assist you in making the most out of TDT.
Tables
At the heart of the Taxonomy Development Tools is a robust internal database designed to streamline the management and curation of taxonomy-related data. Access to this database is facilitated through a user-friendly interface, with tables being a central component.
The default view when loading TDT is a view of all of the the available tables. You can return to this at any time via the table option in the Tables dropdown menu at the top of the interface, which can also be used to select specific tables.
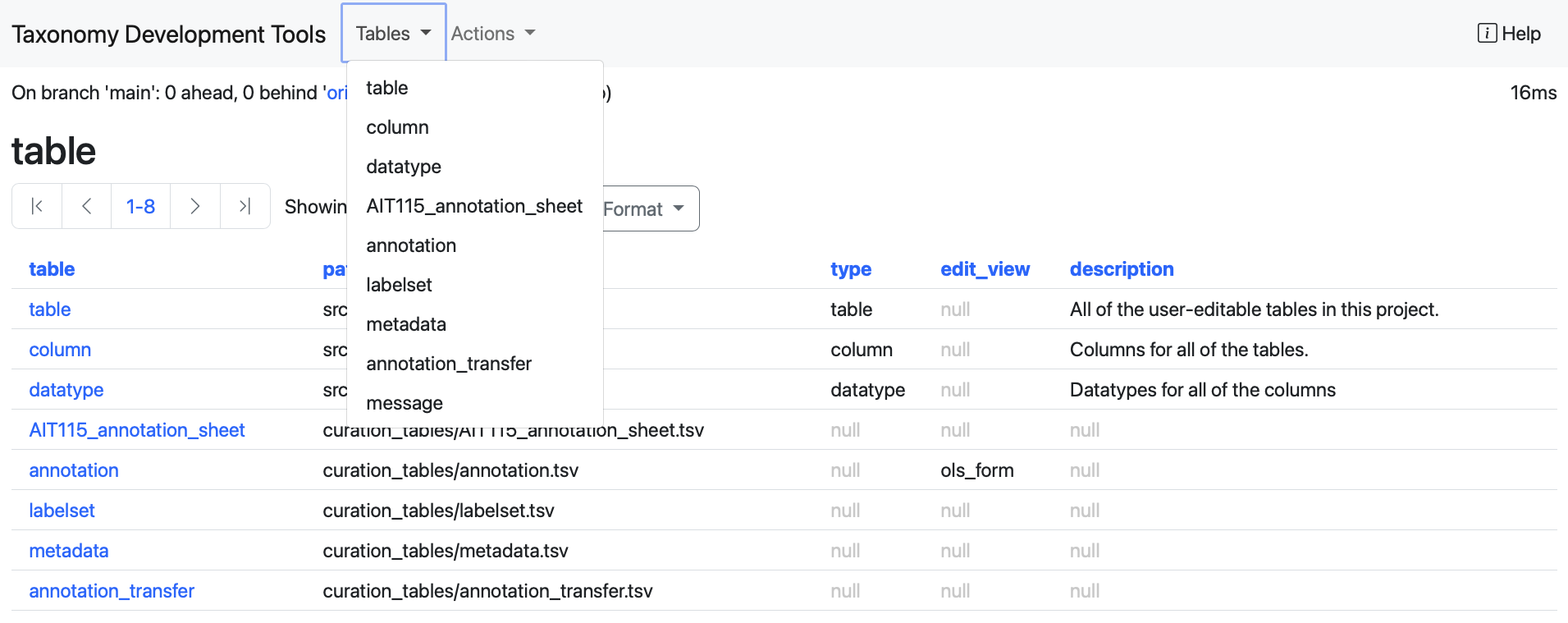
TDT categorizes tables into two main types, system tables and user tables, each serving distinct purposes.
For the purpose of editing taxonomies, our main focus is a set of user tables that reflect taxonomy structure. System tables allow us to inspect the taxonomy schema and to extend it with new columns and datatypes. This is dealt with in the Advanced features section.
User tables
User tables represent the content of the teaxonomy. The main route by with they are are created is when data is uploaded to the TDT using the load_data operation (https://brain-bican.github.io/taxonomy-development-tools/Curation/). This data is formatted according to the Cell Annotation Schema and organized into multiple interrelated tables.
Example: the nhp_basal_ganglia_taxonomy present an annotation table named
AIT115_annotation_sheetfrom this table a series ofuser tablesare generated and displayed in the TDT.
original data table
For taxonomies seeded from an author-provided table, this is present as a reference for the original table. It is not editable an is named accordingo to the {original_table_name}_annotation_sheet.
Example: AIT115_annotation_sheet
metadata
This table contains all the medatadata related to the taxonomy. For full specifications of the metadata properties, look up the cell annotation schema documentation under the section properties.
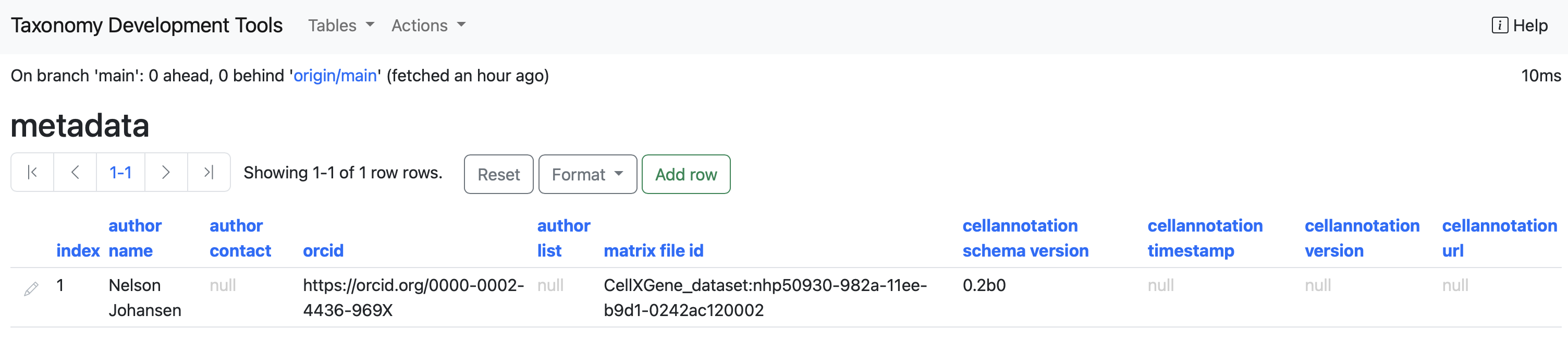
Columns:
author name : The name of the first author of the taxonomy.
author contact : Author's email.
author list : Name of secondary authors.
matrix file ID : A resolvable ID for a cell by gene matrix file.
cellannotation schema version : The version of the cell annotation schema.
cellannotation timestamp : The time (yyyy-mm-dd) of when the cell annotations are published.
cellannotation url : A URL that can be used to resolve the latest published version of this taxonomy (blank if unpublished).
labelset
Annotations are organised into labelsets - keys grouping related annotations. These correspond to cell annotation keys in a cell by gene matrix, for axmple corresponding to obs keys in an AnnData file. This table contains the names and descriptions of labelsets used in the annotation and, optionally, details of the methodology used to acquire those labels. Full specifications of the labelset can be found in the Cell Annotation Schema documentation under the labelsets section.
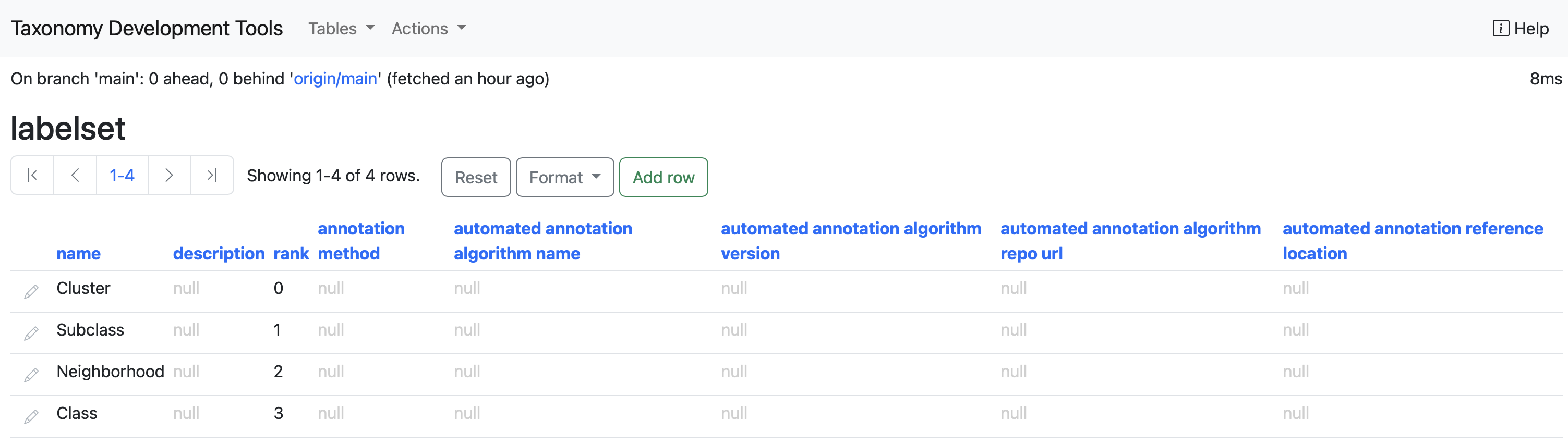
Columns
name : The name of the labelset.
description : Description of the labelset.
rank : The level of granularity of the annotation with 0 being the most specific.
annotation method : The method used for the type of annotation, it can either be algorithmic, manual or both.
automated annotation algorithm name : The name of the algorithm used for the automated annotation.
automated annotation algorithm verision : The version used for the algorithm.
automated annotation algorithm repo url : A resolvable URL of the version control repository of the algorithm used.
automated annotation reference location : A resolvable URL of the source of the data.
annotation:
Stores annotations corresponding to cell types, classes, or states as well as extended metadata about these annotations - including supporting evidence and provenance information. It is designed to be flexible, allowing for additional fields to accommodate user needs or project-specific metadata. In the table, each row corresponds to an annotation of a single cell set - which may be at any level of granularity. Each annotation must belong to a labelset.
The annotation table includes Cell Annotation Standard columns and Project specific columns. Further information on the Cell Annotation Standard columns can be found in the Cell annotation schema documentation under the annotations section.
The Project specific columns are transferred from the original data table. The Project specific columns allow customisation of the annotation table and flexibility over the annotation of the data. Addtional columns can be added via the columns table (see Advanced features)
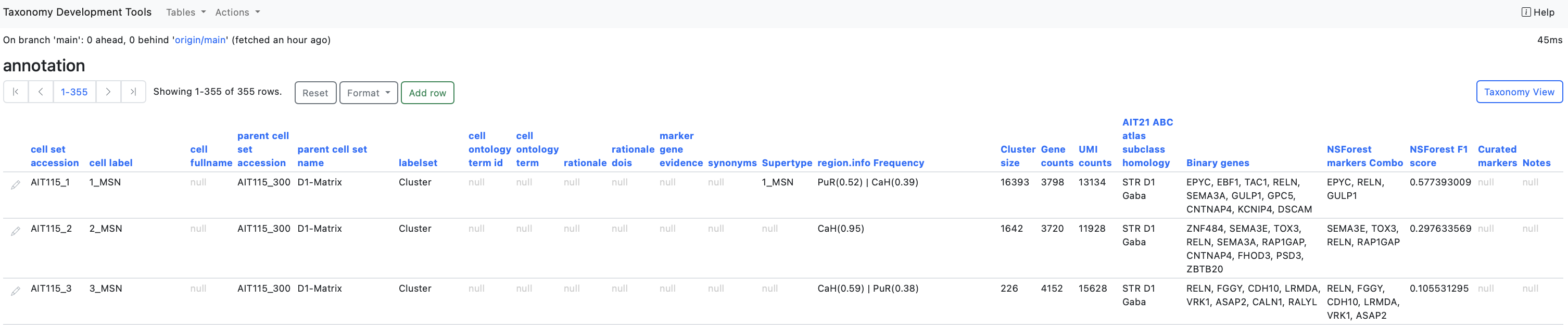
Columns
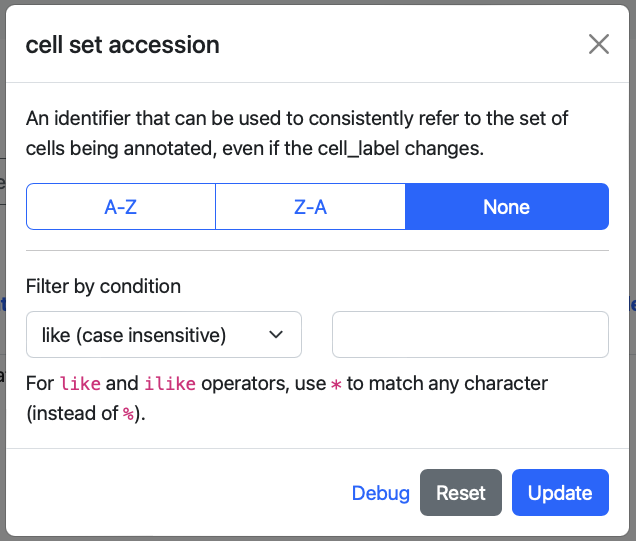
cell set accession : An identifier that can be used to consistently refer to the set of cells being annotated, even if the cell_label changes.
cell label : The cell annotation provided by the author.
cell fullname : Cell labels often use short acronyms or abbreviations which can be opaque to users outside a particular community. This field allows taxonomy editors to spell out these acronyms to avoid ambiguity, e.g. a it might spell out 'interneuron' where the label field uses 'int' or 'medium spiny neuron' where the label field uses 'MSN'.
labelset : Each annotation must belong to one of a set of predefined keys, defined in the labelsets tabe. Each labelset corresponds to a key used to group related annotations in a cell by gene matrix (an obs key in AnnData).
parent cell set accession : The ID of the parent of the set described in this row. This is not editable. It is changed automatically by editing parent cell set name
parent cell set name: The name (label) of the cell set which is the parent of the set described in this row. This row is editable via autocomplete. Editing it changes the parent cell set accession automatically.
cell ontology term id : The ID of the ontology term in the 'cell ontology term' field. This is not editable. It is changed automatically by editing cell ontology term field.
cell ontology term : A term from the cell ontology that most closely matches the cell type/class being annotated in this row.
rationale : A free text field describing the reasons that editors believe the cells in the set to correspond to the cell type referenced in the cell_label, cell_fullname and cell ontology term fields. Editors are encouraged to use this field to cite relevant publications in-line using standard academic citations of the form (Zheng et al., 2020). All references cited SHOULD be listed using DOIs under rationale_dois.
rationale dois : The DOIs of papers supporting the annotation / references in the rationale.
maker gene evidence : List of names of genes whose expression in the cells being annotated is explicitly used as evidence for this cell annotation. Each gene MUST be included in the matrix of the AnnData/Seurat file.
synonyms : Synonyms of the cell label.
Taxonomy view
Annotation tables are equipped to display data in a hierarchical structure, offering a more intuitive understanding of relationships within the taxonomy. To access this perspective, simply click on the Taxonomy View button.
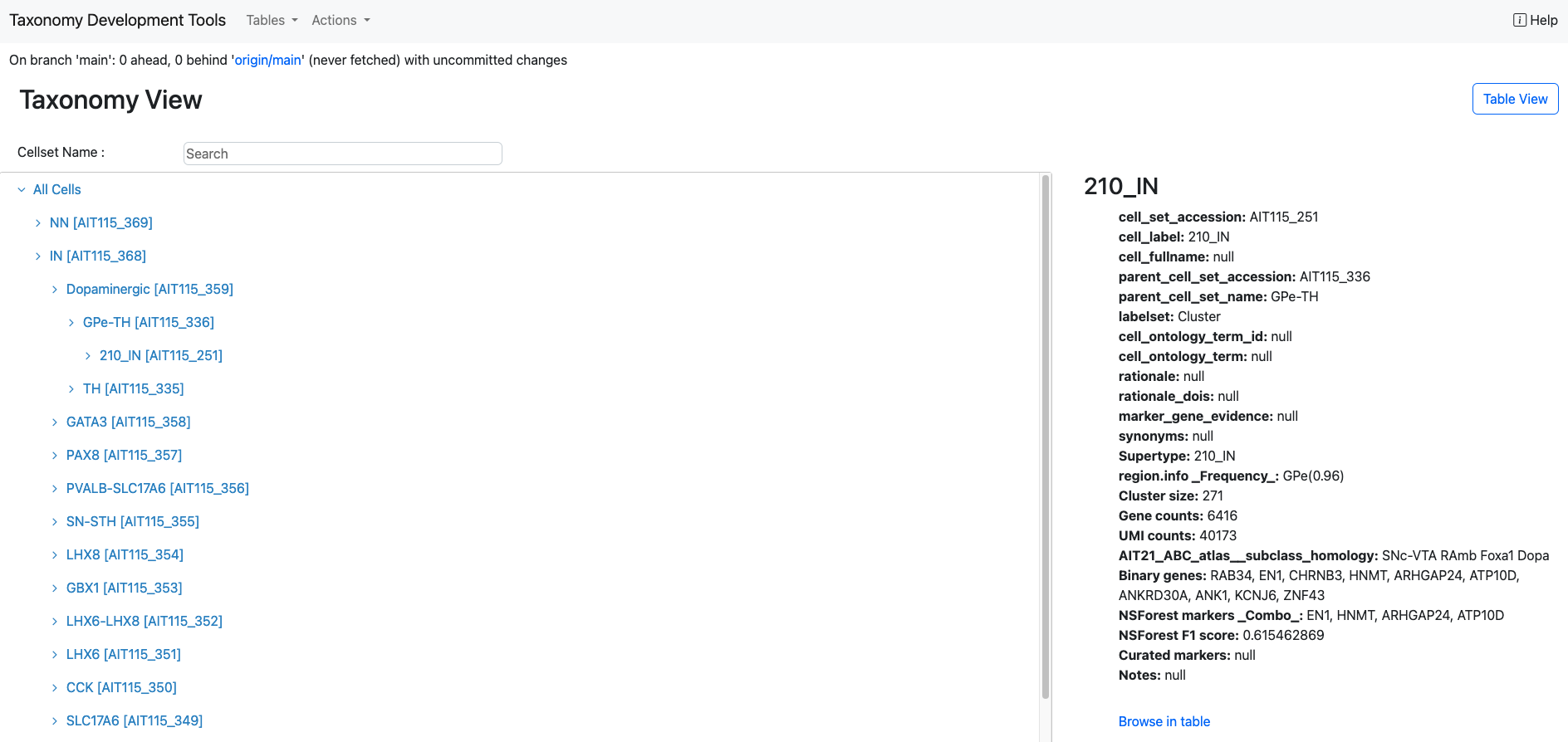
While in the Taxonomy View, users can visually explore the taxonomy's structure in a read-only format, providing a clear overview of the hierarchical relationships. To switch back to a more detailed and interactive tabular format, click the Table View button. Alternatively, users can use Browse in table to jump to the node in the table view.
Search functionality is available in the Taxonomy View, allowing users to quickly locate specific nodes within the taxonomy. By entering a search term in the search bar, users can reveal the term in the taxonomy tree.
Table Management
This section shows how to interact with the Tables within the TDT. In this documentation, the overview of the table management is shown with the annotation table.
Sorting and Filtering Data
By clicking on column names, pop-up widget enabling data-sort and filtering can be activated. This widget allows for the alphabetical sorting of data and the application of conditions to filter data accordingly. Icons next to the column header indicate active sorting or filtering, and clicking the column header again lets you update or clear these parameters.
Reset Table
To reset the sorting and filtering of the table select the Reset button underneath the table name.
Adding New Records
Add row button can be used to add new records to the table. By selecting Add row, a new page will open, start inserting the informations for the new row. The option cell set accession will be auto-filled with a new cell set unique identifier.
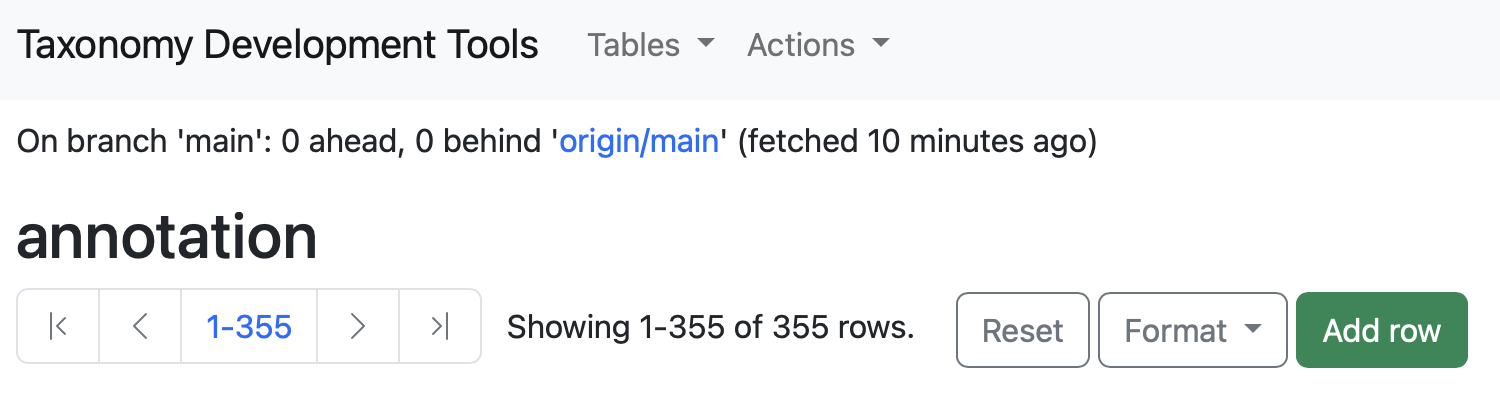
To know what each option represents, hover on the question mark icon near each option.
Editing Existing Data
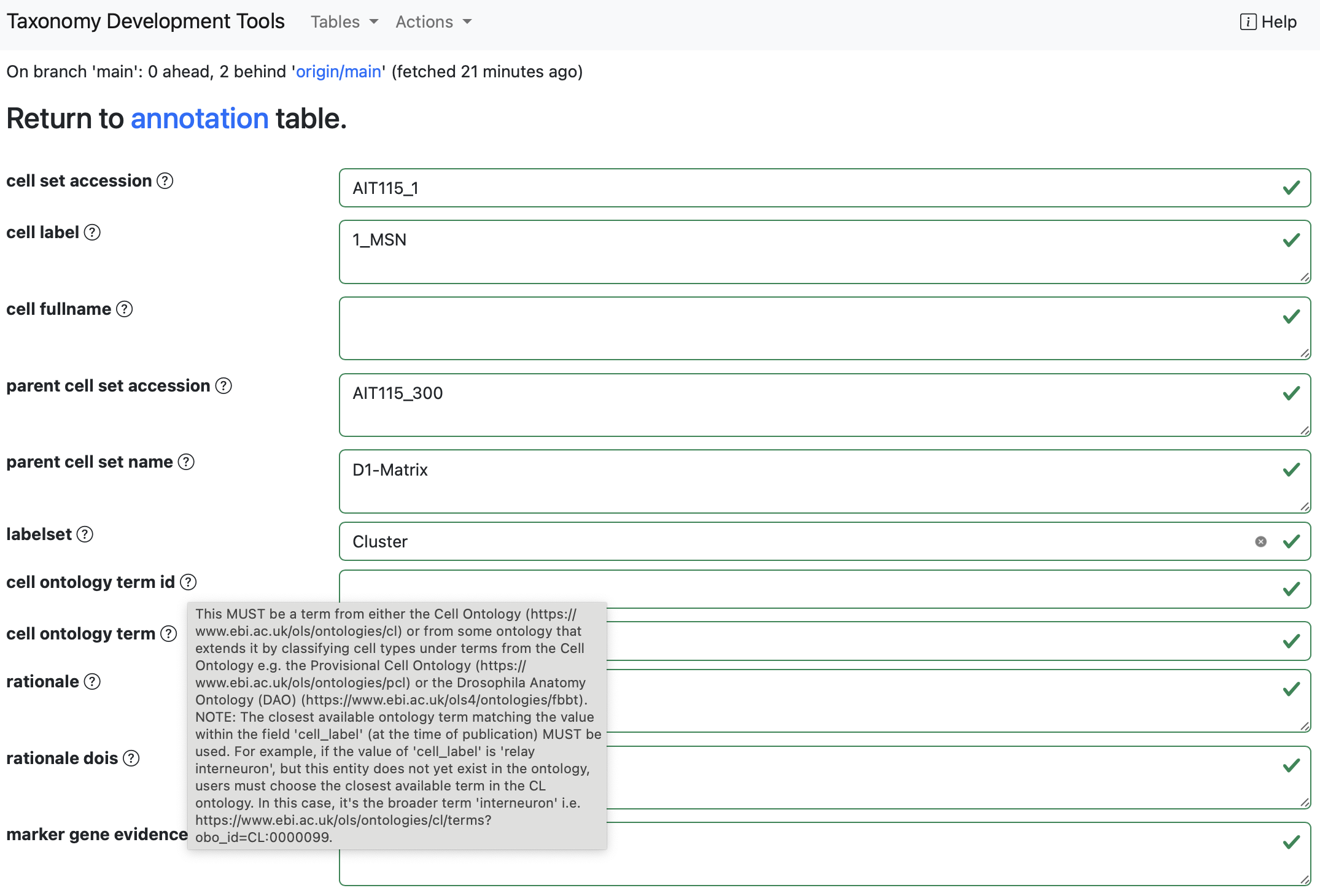
Users can edit records by clicking on the pen icon located at the start of each row. This action directs the user to a data submission form that supports autofill, allowing for efficient data entry. After validating the entered data, users can finalize their edits by clicking the Submit button.
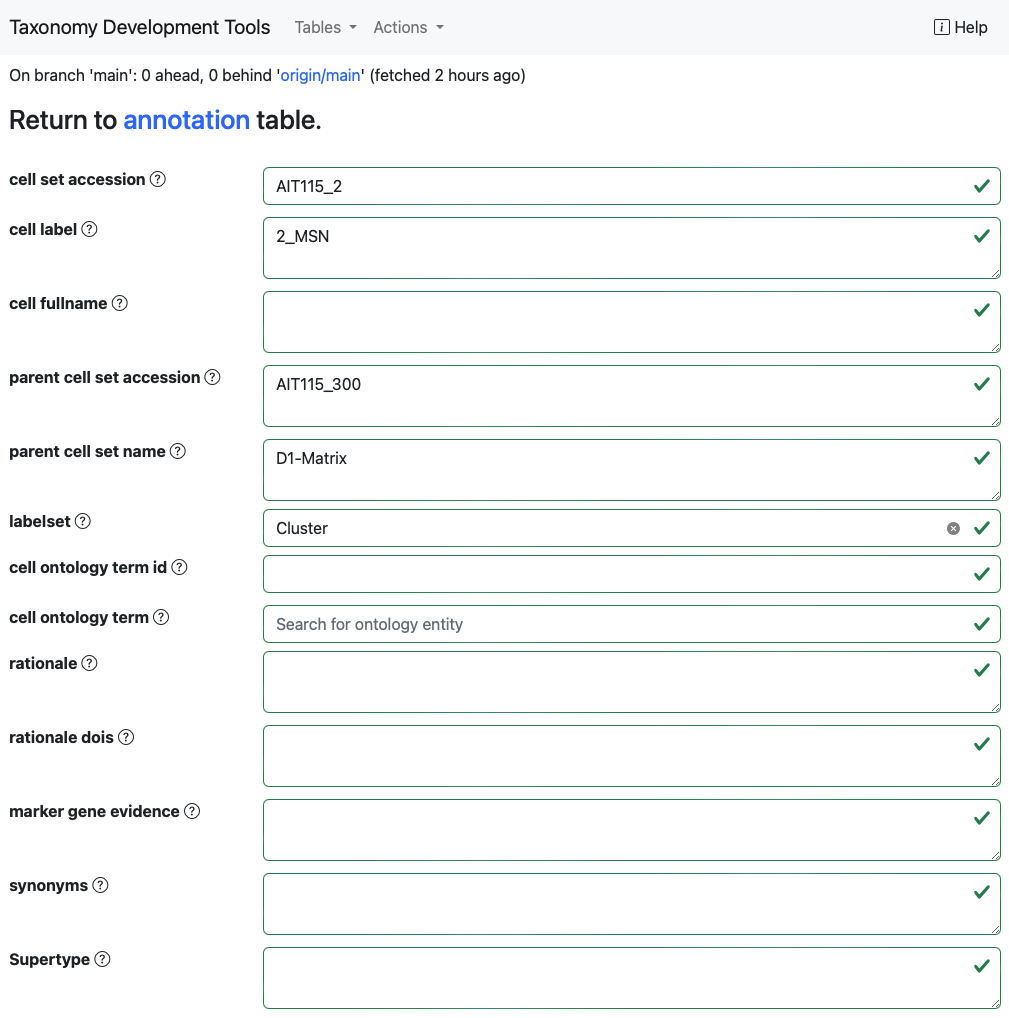
Some options will have a dropdown menu with suggested terms. For exmaple the annotation.labelsetfield has a drop-down list of labelsets defined in thelabelset` table.
Example: In the dataset generated from the
AIT115_annotation_sheet, by selecting the empty box for thelabelsetoption the dropdown options are the labelsetsClass,Cluster,NeighborhoodandSubclass.

Example: When editing the
parent cell set name, suggested terms will appear as a dropdown menu.
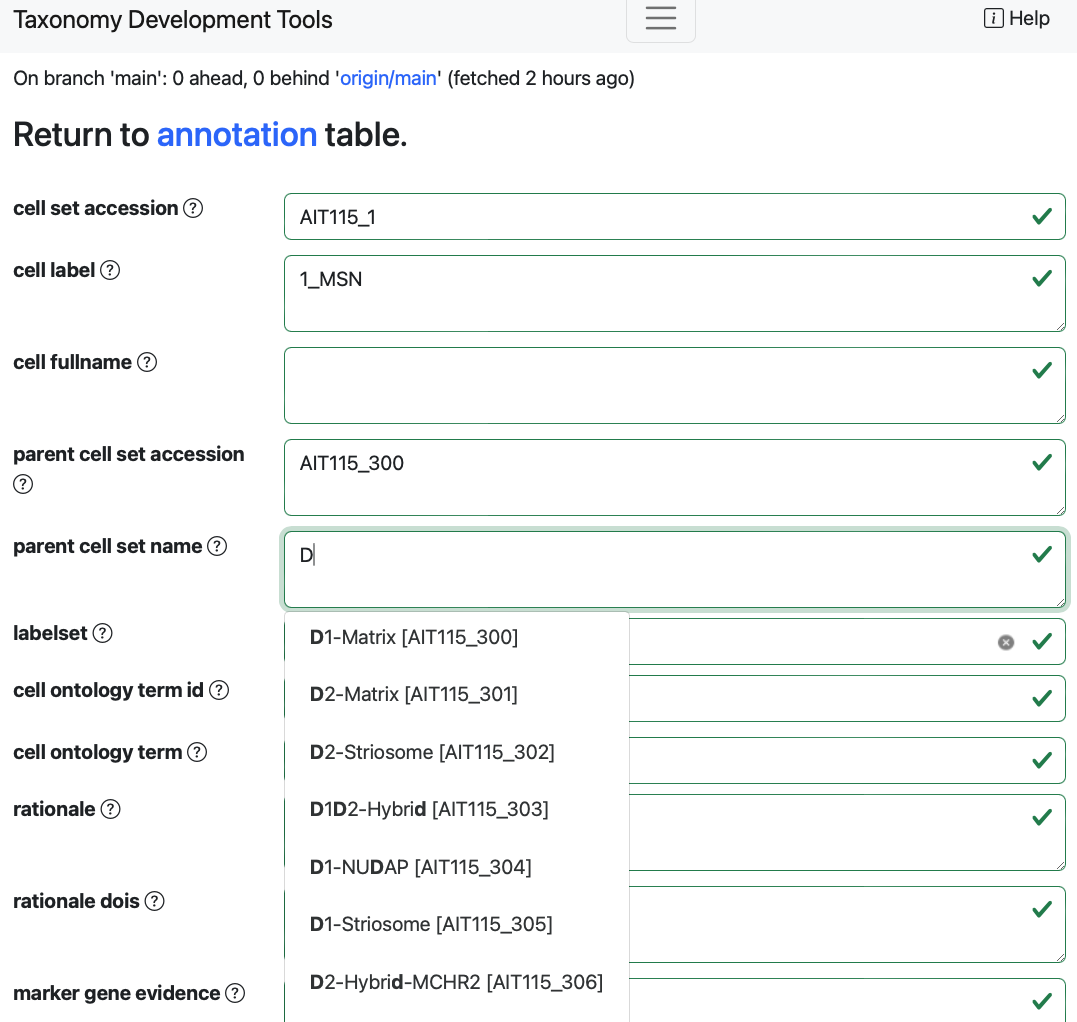
Some options will automatically fill other fields linked to them.
Example: When changing the
parent cell set namefield, theparent cell set accessionfield will also change.

To add a pre-existing Cell Ontology term select the cell ontology term option and start typing the cell term of interest. A dropdown menu will appear with different options for pre-existing ontology terms.
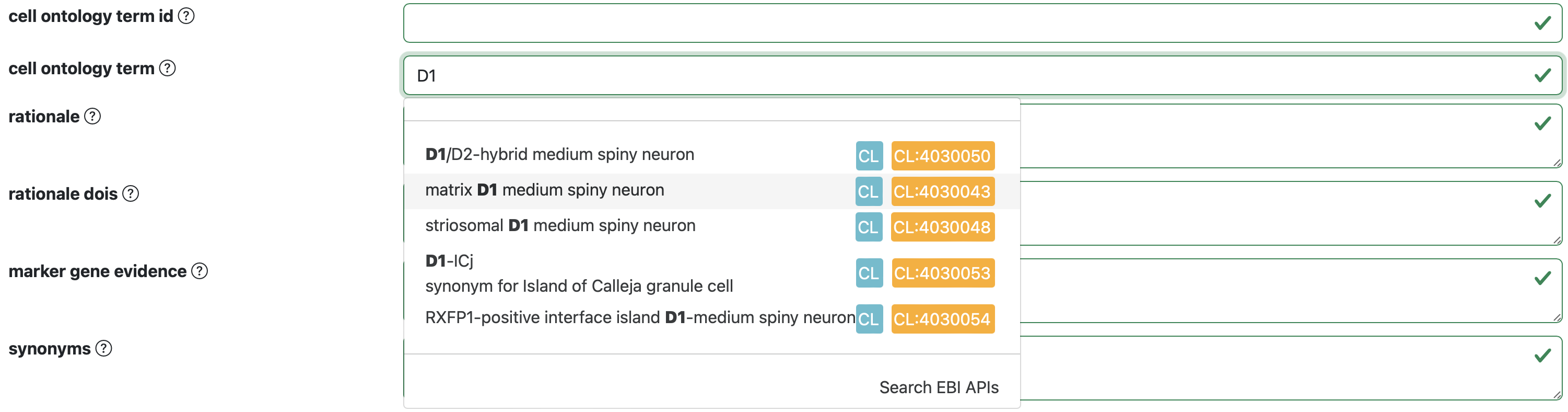
After selecting the cell ontology term of interest, the cell ontology term option and the cell ontology term ID will be autofilled.

Saving existing or new data
To save the modified or newly added row select Submit at the bottom right corner of the page.
The Validate option will ensure that the terms added in each column follow the Cell Annotation Schema and any additional schema specified for this particular taxonomy.

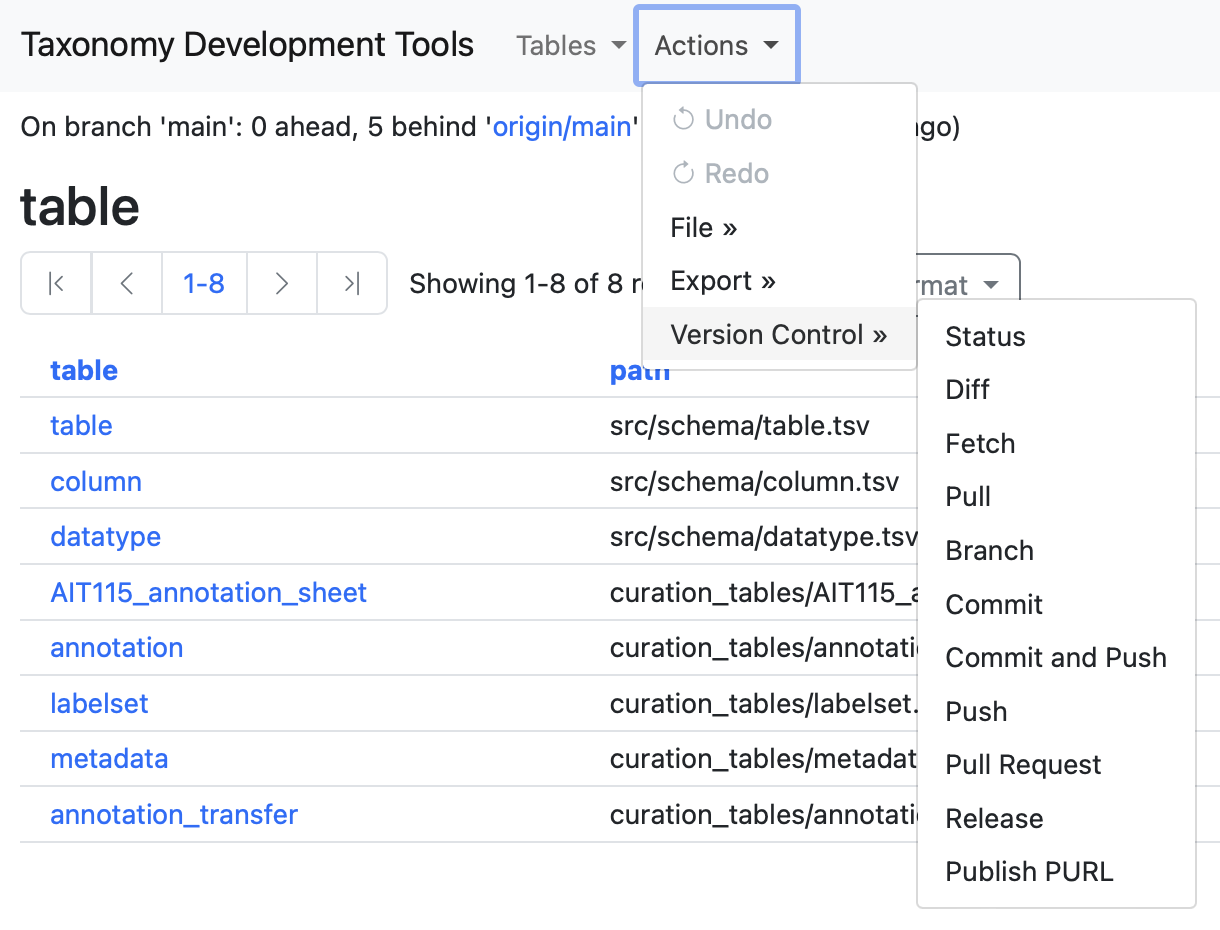
Actions
The following menu includes a series of comands to save and export your data.
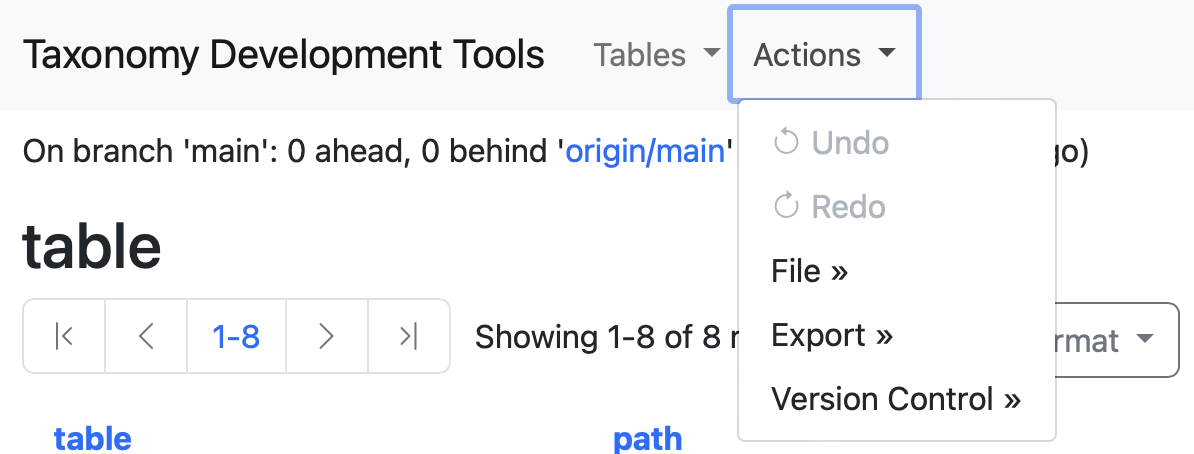
Save
All user modifications are automatically saved to the internal database. However, if user wants to save changes back to the source tsv files located in the project curation_tables/ folder, save action can be utilized.
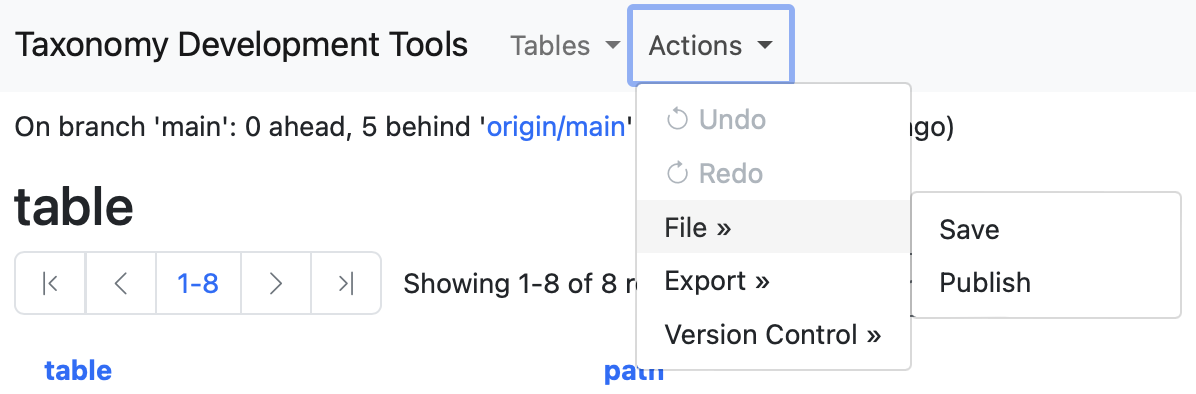
In the Actions menu, select the File dropdown menu. Two options will be displayed save and publish.
The save button allows to save your progress in a syncronised fashion so that other curators of the same taxonomy can see and track the progress. The syncronisation of the saved progress gives the opportunity to multiple users to work collaboratevely on a unified dataset.
The publish button allows to create a public release of the taxonomy on GitHub with a Persistent URL that serves as a resolveable identifier for that release. It is important to note that the accessiblity of a published taxonomy is controlled through GitHub. Editor may choose to publish and share interim versions internally. The Persistent URL serves as a way to permanently referenceand retrieve a version used in analysis.
Export
Under the Actions menu, select the Export drowpdown menu. Two options will be diplayed To CAS and To AnnData.
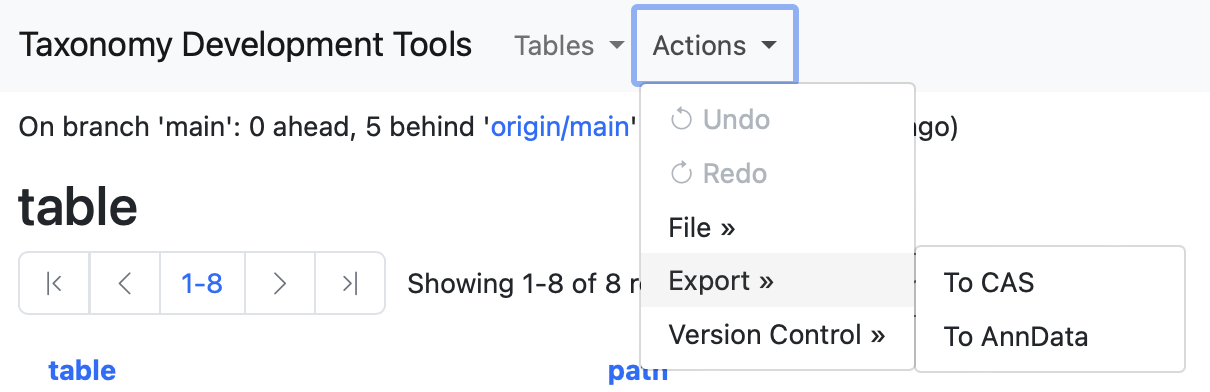
To CAS
This action converts all data into a JSON file formatted according to the Cell Annotation Schema located in the project's root directory.
To AnnData
Apply your modifications directly to the associated AnnData file for the taxonomy.
The metadata table contains a matrix_file_id column, representing a resolvable ID for a cell-by-gene matrix file, formatted as namespace:accession. For example, CellXGene_dataset:8e10f1c4-8e98-41e5-b65f-8cd89a887122 (Please refer to the cell-annotation-schema registry for a list of supported namespaces.)
Upon initial launch, TDT creates a tdt_datasets directory in the user home folder. It attempts to resolve all matrix_file_id references within this directory. The system searches for the corresponding file (e.g., $HOME/tdt_datasets/8e10f1c4-8e98-41e5-b65f-8cd89a887122.h5ad) and then initiates a cas-tools flatten operation. This process, which may vary in duration depending on the taxonomy's size, results in a new AnnData file (/tdt_datasets/$$TAXONOMY_ID$$.h5ad) incorporating the user's annotations.
Advanced options
System tables:
System tables allow us to inspect the taxonomy schema and to extend it with new columns and datatypes.
table: This table lists all the tables present in the TDT and it appears in the default page of the TDT.
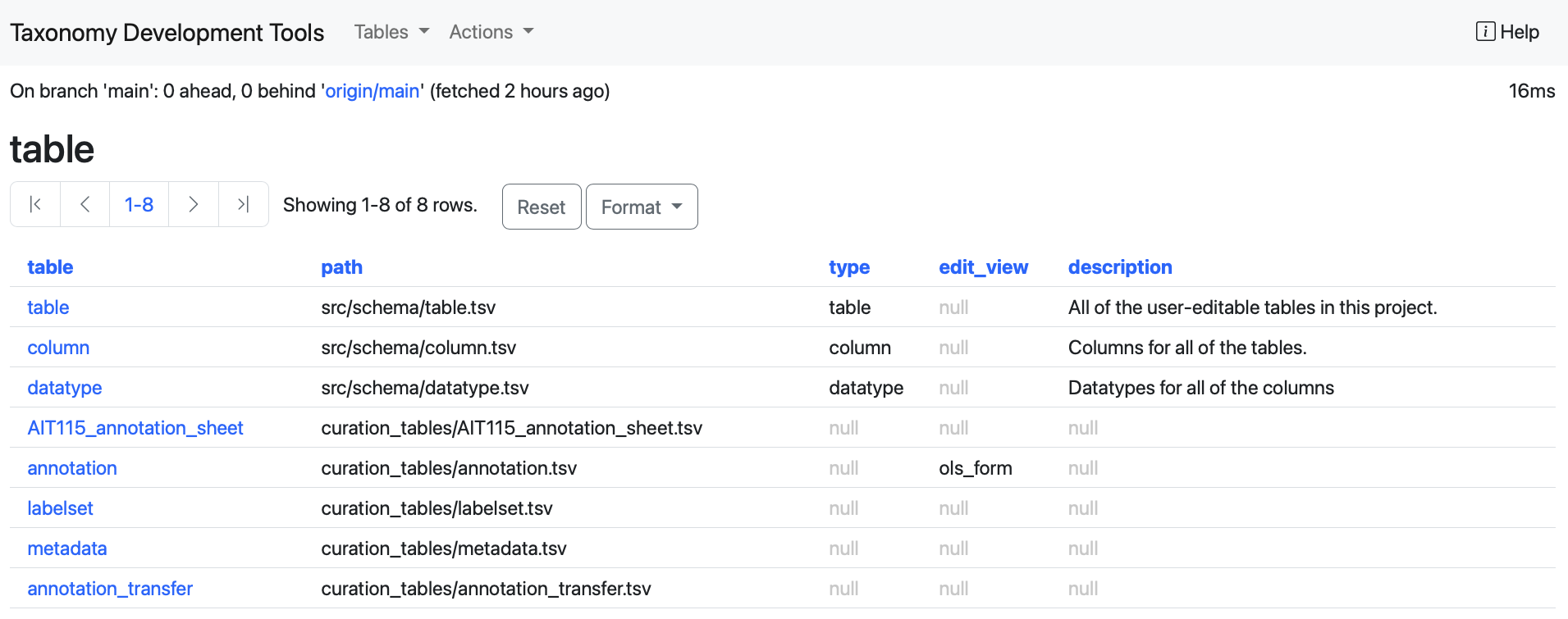
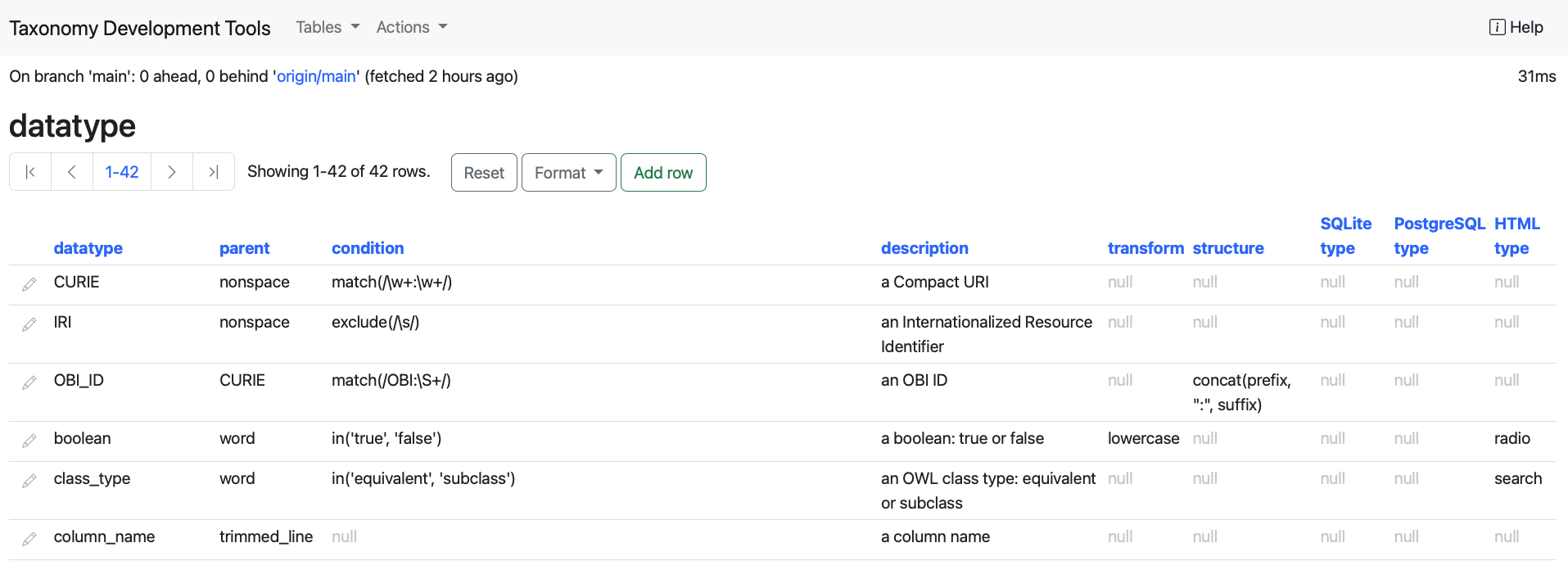
datatype: this table shows all the datatype columns present in each table.
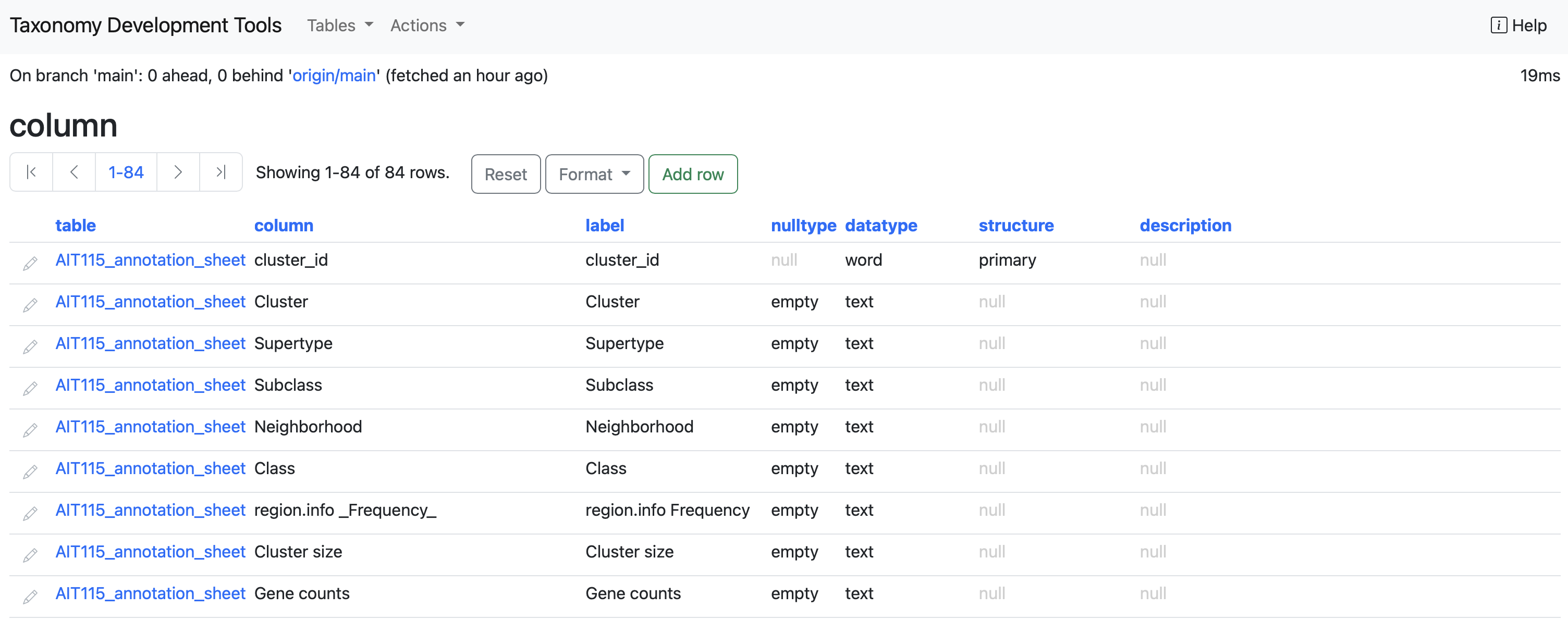
column: This table contains all the columns present in each table.
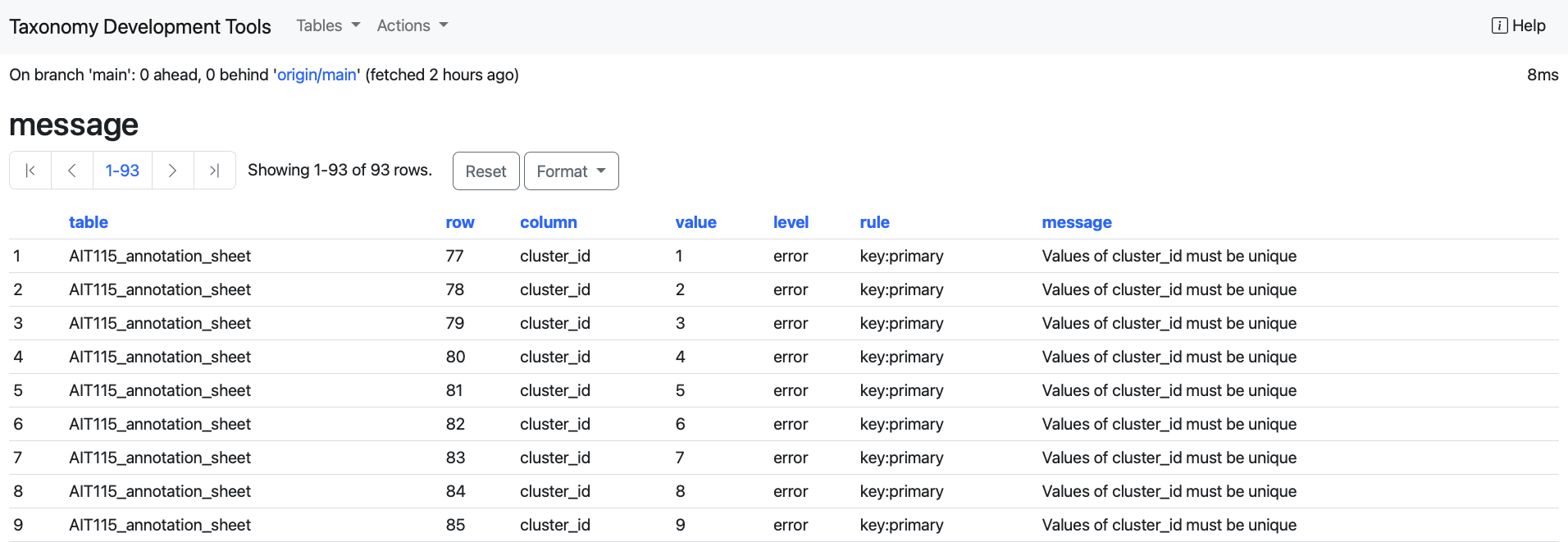
message: This table contains all validation error messages.
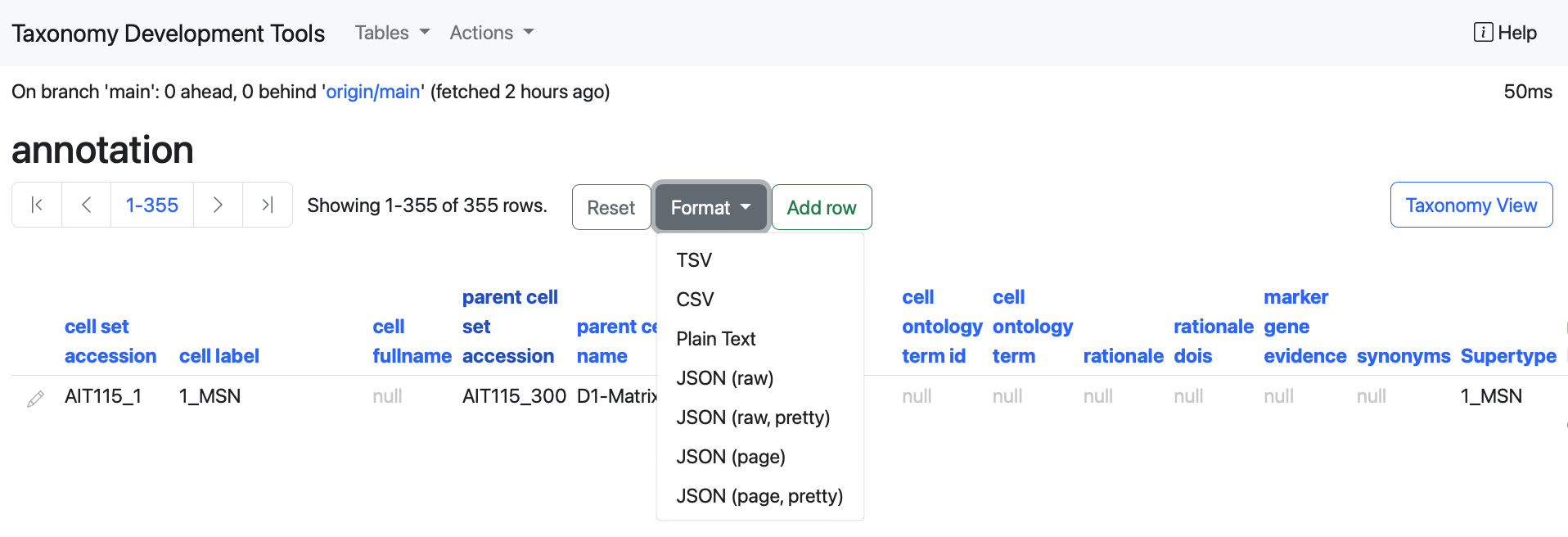
Change Table Format
The table format can be changed by selecting the Format button underneath the table name. This will open a dropdown menu with a series of options to display the data in different format of a TSV or a CSV table; Plain Text or in a Json raw or page format (for more information about Json format have a look at additional resources). In addition to the Json raw or page option, the options Json(raw, pretty) and Json(page, pretty) are available to display the data in a Json format that is easier to read.
### Version Control
Under the Action dropdown menu, a complete set of Git version control options are available. The verson control menu is intended for comunities of users who are comfortable working with GitHub.